Research
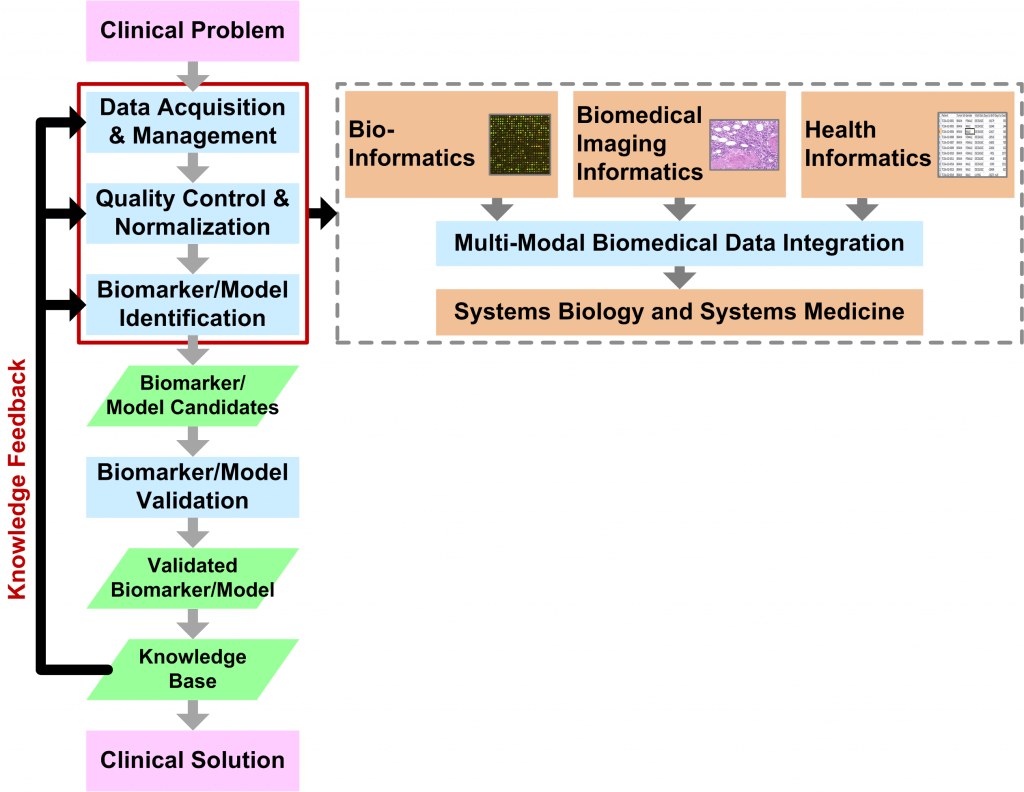
Bio-MIBLab research primarily focuses on Translational Biomedical Informatics (Figure) with the goal of developing software applications and algorithms that solve real-world clinical problems. Such applications and algorithms are essential for personalized, predictive, and preventive medicine. The emergence of “Big Data” in the areas of bioinformatics (e.g., massively parallel genomic sequencing and mass spectrometry), biomedical imaging informatics (e.g., whole-slide histopathological imaging), and health informatics (e.g., physiological patient data and electronic health records) have changed the way that we analyze and visualize data. In short, the enormous data volume is an immediate challenge that requires novel solutions in terms of algorithms (e.g., data integration, distributed computing, and visualization) as well as technology (e.g., data transfer and storage).
The Translational Biomedical Informatics Pipeline provides methodological guidelines for producing solutions in the form of biomarkers and biological models. Validated biomarkers and models make up the knowledge-base, which, in turn, guides the core analytical steps in an iterative feedback process. These core analytical steps include: (1) Data Acquisition/Management, (2) Quality Control/Normalization, and (3) Biomarker/Model Identification. These analytical steps are independently applicable to four core research areas covering different data scales:
- Bioinformatics
- Biomedical Imaging Informatics
- Health Informatics
- Systems Biology and Systems Medicine
Systems Biology and Systems Medicine is a broad application of methods from the other core research areas and encompasses Multi-Modal Biomedical Data Integration, which, in itself, is a rich area of research in algorithms and data analytics.