Predictive Modeling
Gene expression-based prediction modeling
- Common practices for microarray-based prediction models (MAQC-II study)
- KNN models for microarray-based clinical outcome prediction (MAQC-II study)
- omniClassifier: Big Data prediction modeling
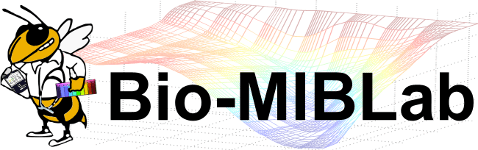
The MAQC-II Project: A Comprehensive Study of Common Practices for the Development and Validation of Microarray-Based Predictive Models
Gene expression data from microarrays are being applied to predict preclinical and clinical endpoints, but the reliability of these predictions has not been established. In the MAQC-II project, 36 independent teams analyzed six microarray data sets to generate predictive models for classifying a sample with respect to one of 13 endpoints indicative of lung or liver toxicity in rodents, or of breast cancer, multiple myeloma or neuroblastoma in humans. In total, more than 30,000 models were built using many combinations of analytical methods. The teams generated predictive models without knowing the biological meaning of some of the endpoints and, to mimic clinical reality, tested the models on data that had not been used for training. We found that model performance depended largely on the endpoint and team proficiency and that different approaches generated models of similar performance. The conclusions and recommendations from MAQC-II are useful for regulatory agencies, study committees and independent investigators that evaluate methods for global gene expression analysis.
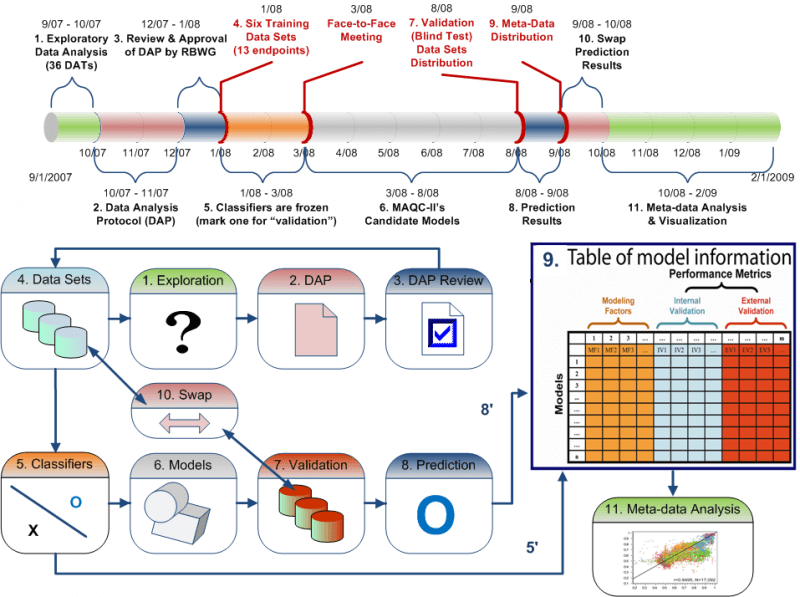
K-Nearest Neighbor Models for Microarray Gene Expression Analysis and Clinical Outcome Prediction
In the clinical application of genomic data analysis and modeling, a number of factors contribute to the performance of disease classification and clinical outcome prediction. This study focused on the k-nearest neighbor (KNN) modeling strategy and its clinical use. Although KNN is simple and clinically appealing, large performance variations were found among experienced data analysis teams in the MicroArray Quality Control Phase II (MAQC-II) project. For clinical end points and controls from breast cancer, neuroblastoma and multiple myeloma, we systematically generated 463,320 KNN models by varying feature ranking method, number of features, distance metric, number of neighbors, vote weighting and decision threshold. We identified factors that contribute to the MAQC-II project performance variation, and validated a KNN data analysis protocol using a newly generated clinical data set with 478 neuroblastoma patients. We interpreted the biological and practical significance of the derived KNN models, and compared their performance with existing clinical factors.
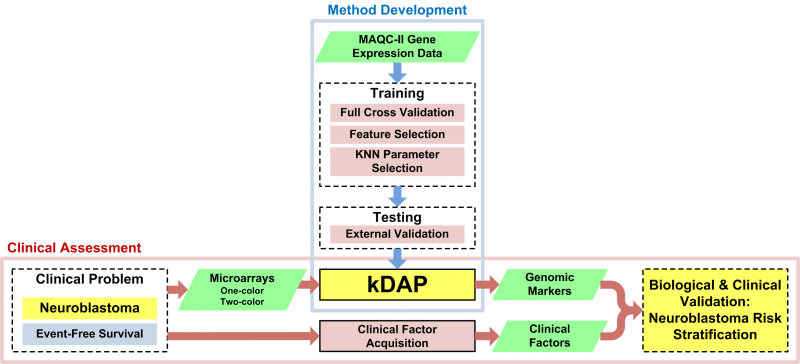
omniClassifier: A Desktop Grid Computing System for Big Data Prediction Modeling
Robust prediction models are important for numerous science, engineering, and biomedical applications. However, best-practice procedures for optimizing prediction models can be computationally complex, especially when choosing models from among hundreds or thousands of parameter choices. Computational complexity has further increased with the growth of data in these fields, concurrent with the era of “Big Data“. Grid computing is a potential solution to the computational challenges of Big Data. Desktop grid computing, which uses idle CPU cycles of commodity desktop machines, coupled with commercial cloud computing resources can enable research labs to gain easier and more cost effective access to vast computing resources. We developed omniClassifier, a multi-purpose prediction modeling application that provides researchers with a tool for conducting machine learning research within the guidelines of recommended best-practices. omniClassifier is implemented as a desktop grid computing system using the Berkeley Open Infrastructure for Network Computing (BOINC) middleware. We used various gene expression datasets to demonstrate the potential scalability of omniClassifier for efficient and robust Big Data prediction modeling.
http://omniclassifier.bme.gatech.edu/