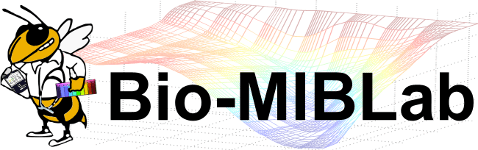
WSI Quality Control
Algorithms and applications that we have developed for the histopathological image quality control can be divided into the following categories:
- Artifact detection and elimination in histopathological whole-slide images
- Evaluation of batch effect normalization for histopathological images
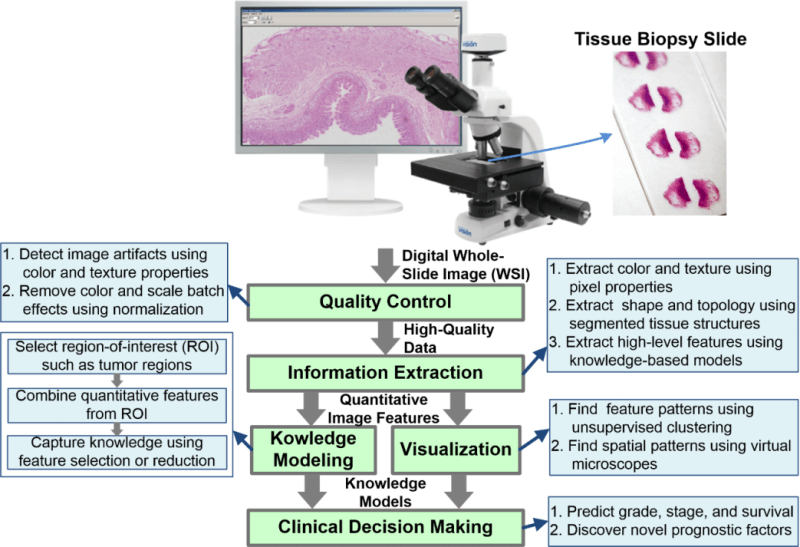
Artifact Detection and Elimination in WSIs
Analysis of tissue biopsy whole-slide images (WSIs) depends on effective detection and elimination of image artifacts. We developed a novel method to detect tissue-fold artifacts in histopathological WSIs. We also studied the effect of tissue folds on image features and prediction models. We used WSIs of samples from two cancer endpoints–kidney clear cell carcinoma (KiCa) and ovarian serous adenocarcinoma (OvCa)–publicly available from The Cancer Genome Atlas. We detected tissue folds in low-resolution WSIs using color properties and two adaptive connectivity-based thresholds. We optimized and validated our tissue-fold detection method using 105 manually annotated WSIs from both cancer endpoints. In addition to detecting tissue folds, we extracted 461 image features from the high-resolution WSIs for all samples. We used the rank-sum test to find image features that were statistically different among features extracted from the same set of WSIs with and without folds. We then used features that were affected by tissue folds to develop models for predicting cancer grades.
When compared to the ground truth, our method detected tissue folds in KiCa with 0.50 adjusted Rand index (ARI), 0.77 average true rate (ATR), 0.55 true positive rate (TPR), and 0.98 true negative rate (TNR); and in OvCa with 0.40 ARI, 0.73 ATR, 0.47 TPR, and 0.98 TNR. Compared to two other methods, our method was more accurate in terms of ARI and ATR. We found that 53 and 30 image features were significantly affected by the presence of tissue-fold artifacts (detected using our method) in OvCa and KiCa, respectively. After eliminating tissue folds, the performance of cancer-grade prediction models improved by 5% and 1% in OvCa and KiCa, respectively. Our proposed connectivity-based method was more effective in detecting tissue folds compared to other methods. Reducing tissue-fold artifacts will increase the performance of cancer-grade prediction models.
Evaluation of Batch Effect Normalization Methods for WSIs
Researchers have developed computer-aided decision support systems for translational medicine that aim to objectively and efficiently diagnose cancer using histopathological images. However, the performance of such systems is confounded by non-biological experimental variations or “batch effects” that can commonly occur in histopathological data, especially when images are acquired using different imaging devices and patient samples. This is even more problematic in large-scale studies in which cross-laboratory sharing of large volumes of data is necessary. Batch effects can change quantitative morphological image features and decrease prediction performance. Using four batches of renal tumor images, we compared one image-level and five feature-level batch effect removal methods. Principal component variation analysis (PCVA) showed that batch is a large source of variance in image features. Our analysis revealed that feature-level normalization methods reduced batch-contributed variance to almost zero. Moreover, feature-level normalization, especially ComBatN, improved cross-batch and combined-batch prediction performance. Compared to no normalization, ComBatN improved performance in 83% and 90% of cross-batch and combined-batch prediction models, respectively.